PUBLICATIONS
|
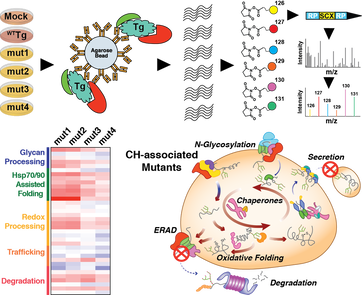
57. Davies JP, Plate L. The glycoprotein quality control factor Malectin promotes coronavirus replication and viral protein biogenesis. [bioRxiv preprint]. 2024 Jun.; doi: 10.1101/2024.06.02.597051
56. Egly CL, Do TQ, McDonald EF, Barny L, Knollmann B, Plate L. The proteostasis interactomes of trafficking-deficient variants of the voltage-gated potassium channel KV11.1 associated with Long QT Syndrome. Journal of Biological Chemistry. 2024 [June 12, in press]; doi: 10.1016/j.jbc.2024.107465 |
55. Kim M, Plate L. Cystic Fibrosis Modulator Therapies: Bridging Insights from CF to other Membrane Protein Misfolding Diseases. Israel Journal of Chemistry. [in press] Jan. 24, 2024. doi: 10.1002/ijch.202300152
|
54. McDonald EF, Oliver KE, Schlebach JP, Meiler J, Plate L. Benchmarking AlphaMissense Pathogenicity Predictions Against Cystic Fibrosis Variants. PLOS ONE. 2024 Jan. 25; doi: 10.1371/journal.pone.0297560
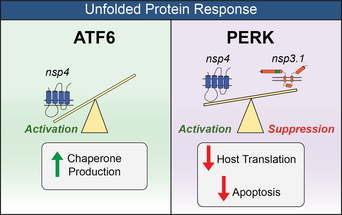
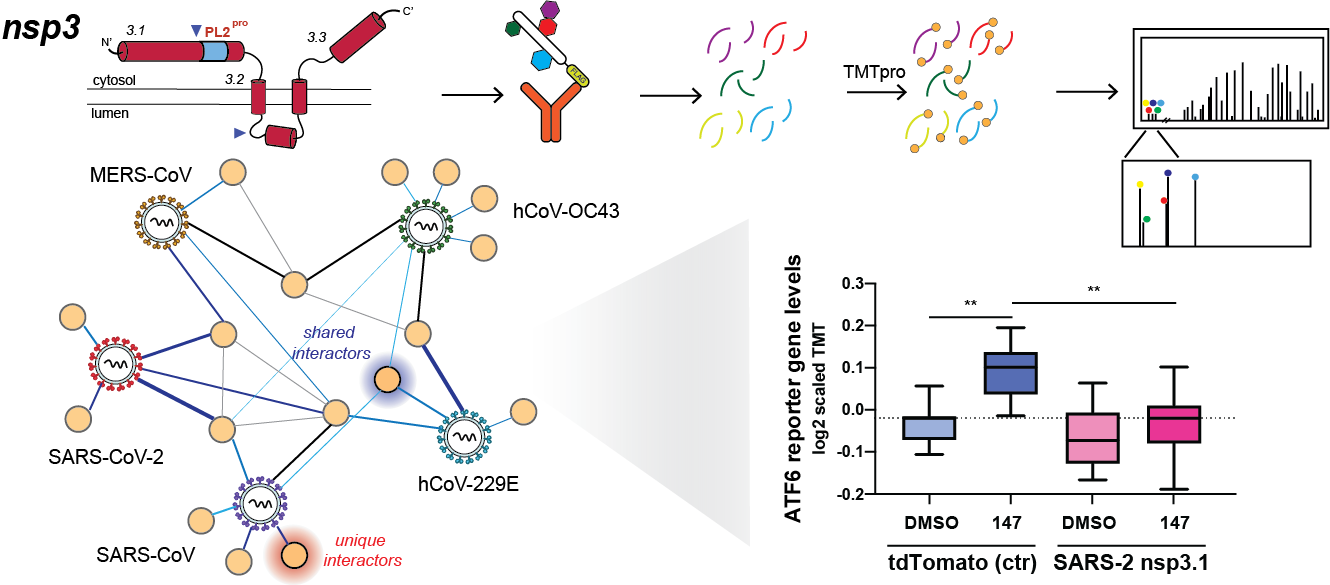
53. Davies JP, Sivadas A, Keller KR, Roman BK, Wojcikiewicz RJH, Plate L. Expression of SARS-CoV-2 Nonstructural Proteins 3 and 4 Can Tune the Unfolded Protein Response in Cell Culture. Journal of Proteome Research. 2024 (23): 356; doi: 10.1021/acs.jproteome.3c00600 |
|
52. Sivadas A, McDonald EF, Shuster SO, Davis CM, Plate L. Site-Specific Crosslinking Reveals Phosphofructokinase-L Inhibition Drives Self-Assembly and Attenuation of Protein Interactions. Advances in Biological Regulation. 2023 (90): 100987. doi: 10.1016/j.jbior.2023.100987doi.org/10.1016/j.jbior.2023.100987
51. McDonald EF, Meiler J, Plate L. CFTR Folding: From Structure and Proteostasis to Cystic Fibrosis Personalized Medicine. ACS Chemical Biology. 2023 (18): 2128; doi: 10.1021/acschembio.3c00310 |
|
50. Kim M, McDonald EF, Sabusap CMP, Timalsina B, Joshi D, Hong JS, Rab A, Sorscher EJ, Plate L. Elexacaftor/VX-445-mediated CFTR interactome remodeling reveals differential correction driven by mutation-specific translational dynamics. Journal of Biological Chemistry. 2023 (229): 10524;
doi: 10.1016/j.jbc.2023.105242 Article highlighted on the Nov. 2023 Cover of JBC 49. Hazari Y, Urra H, Garcia Lopez VA, Diaz J, Tamburini G, Milani M, Pihan P, Durand S, Aprahamia F, Baxter R, Huang M, Dong XC, Vihinen H, Batista-Gonzales A, Godoy P, Criollo A, Ratziu V, Foufelle F, Hengstler JG, Jokitalo E, Bailly-Maitre B, Maiers JL, Plate L, Kroemer G, Hetz C. The endoplasmic reticulum stress sensor IRE1 regulates collagen secretion through the enforcement of the proteostasis factor P4HB/PDIA1 contributing to liver damage and fibrosis. [bioRxiv preprint]. 2023 May; doi: 10.1101/2023.05.02.538835 |
48. Carmody P, Roushar FJ, Tedman A, Herwig M, Kim M, McDonald EF, Wong-Roushar J, Poirier JL, Zelt N, Pockrass B, McKee AG, Kuntz CP, Plate L, Penn WP, Schlebach JP. Ribosomal Frameshifting Selectively Modulates the Biosynthesis, Assembly, and Function of a Misfolded CFTR Variant. [bioRxiv preprint]. 2023 May; doi: 10.1101/2023.05.02.539166
47. Han D, Schaffner SH, Davies JP, Benton ML, Plate L, Nordman JT. BRWD3 promotes KDM5 degradation to maintain H3K4 methylation levels. PNAS. 2023 (120): e2305092120; doi: 10.1073/pnas.2305092120
46. Duran-Aniotz C, Poblete N, Rivera-Krstulovic C, Ardiles ÁO, Díaz ML, Tamburini G, Sabusap CMP, Gerakis Y, Cabral Miranda F, Diaz J, Fuentealba M, Arriagada D, Muñoz E, Espinoza S, Martinez G, Quiroz G, Medinas DB, Contreras D, Piña R, Lourenco MV, Ribeiro FC, Ferreira ST, Rozas C, Morales B, Plate L, Gonzalez-Billault C, Palacios AG, Hetz C. The unfolded protein response transcription factor XBP1s ameliorates Alzheimer's disease by improving synaptic function and proteostasis. Molecular Therapy. 2023 (23): 00161. doi: 10.1016/j.ymthe.2023.03.028
45. McDonald EF, Jones T, Plate L, Meiler J, Gulsevin A. Benchmarking AlphaFold2 on peptide structure prediction. Structure. 2023 (31): 1 doi: 10.1016/j.str.2022.11.012
44. McKee AG, McDonald EF, Penn WP, Kuntz CP, Noguera K, Chamness LM, Roushar FJ, Meiler J, Oliver KE, Plate L, Schlebach JP. General trends in the effects of VX-661 and VX-445 on the plasma membrane expression of clinical CFTR variants. Cell Chemical Biology. 2023 (30): 632; doi: 10.1016/j.chembiol.2023.05.001
|
43. Tigar R, Davies JP, Plate L, Nordman, JT. The histone chaperone NASP maintains H3-H4 reservoirs in the early Drosophila embryo. PLOS Genetics. 2023 (19): e1010682; doi: 10.1371/journal.pgen.1010682
42. Wright MT, Timalsina B, Garcia Lopez VA, Plate L. Time-Resolved Interactome Profiling Deconvolutes Secretory Protein Quality Control Dynamics. [bioRxiv preprint]. 2022 Sept.; doi: 10.1101/2022.09.04.506558 |
|
41. Cabral-Miranda F, Tamburini G, Martinez G, Medinas D, Gerakis Y, Miedema T, Duran-Aniotz C, Ardiles AO, Gonzalez C, Sabusap C, Bermedo-Garcia F, Adamson S, Vitangcol K, Huerta H, Zhang X, Nakamura T, Pablo Sardi S, Lipton SA, Kenedy BK, Cárdenas J, Palacios AG, Plate L, Henriquez J, Hetz C. Unfolded protein response IRE1/XBP1 signaling is required for healthy mammalian brain aging. EMBO Journal 2022 (41):e111952; doi: 10.15252/embj.2022111952.
Article highlighted on the Nov. 17 2022 Cover of EMBO Journal |
|
40. Sanders AL, Hermanson JN, Samuels DC, Plate L, Sanders CR. Compendium of Proteins Containing Segments That Exhibit Zero-Tolerance to Amino Acid Variation in Humans. Protein Sciences. 2022 (31): e4408; doi: 10.1002/pro.4408
Article highlighted on Vanderbilt Basic Sciences News. 39. McDonald EF, Sabusap CMP, Kim M, Plate L. Distinct proteostasis states drive pharmacologic chaperone susceptibility for Cystic Fibrosis Transmembrane Conductance Regulator misfolding mutants. Molecular Biology of the Cell. 2022 (33): ar62; doi: 10.1091/mbc.E21-11-0578 Article highlighted on the June 2022 cover of Molecular Biology of the Cell. 38. McDonald EF, Woods H, Smith S, Kim M, Schroeder CT, Plate L, Meiler J. Structural comparative modeling of multi-domain F508del CFTR. Biomolecules 22 (12): 471; doi: 10.3390/biom12030471 |
37. Munden A, Wright MT, Han H, Tirgar R, Plate L, Nordman JT. Identification of replication fork-associated proteins in Drosophila embryos and cultured cells using iPOND coupled to quantitative mass spectrometry. Scientific Reports. 2022 (12): 6309 doi: 10.1038/s41598-022-10821-9
36. Bilches Medinas D, Malik S, Yıldız-Bölükbaşı E, Borgonovo J, Saaranen MJ, Urra H, Pulgar E, Afzal M, Contreras D, Wright MT, Bodaleo F, Quiroz G, Rozas P, Mumtaz S, Díaz R,
Rozas C, Cabral-Miranda F, Piña R, Valenzuela V, Uyan O, Reardon C, Woehlbier U, Brown RH, Sena-Esteves M, Gonzalez-Billault, C, Morales B, Plate L, Ruddock LW, Concha ML, Hetz C, Tolun A. Mutation in protein disulfide isomerase A3 causes neurodevelopmental defects by disturbing endoplasmic reticulum proteostasis. EMBO Journal. 2021: e105531; doi: 10.15252/embj.2020105531
Rozas C, Cabral-Miranda F, Piña R, Valenzuela V, Uyan O, Reardon C, Woehlbier U, Brown RH, Sena-Esteves M, Gonzalez-Billault, C, Morales B, Plate L, Ruddock LW, Concha ML, Hetz C, Tolun A. Mutation in protein disulfide isomerase A3 causes neurodevelopmental defects by disturbing endoplasmic reticulum proteostasis. EMBO Journal. 2021: e105531; doi: 10.15252/embj.2020105531
|
35. Reisman BJ, Guo H, Ramsey HE, Wright MT, Reinfeld BI, Ferrell PB, Sulikowski GA, Rathmell WK, Savona MR, Plate L, Rubinstein JL, Bachmann BO. Apoptolidin family glycomacrolides target leukemia through inhibition of ATP synthase. Nature Chemical Biology. 2022 (18): 360; doi: 10.1038/s41589-021-00900-9
Article highlighted in NCB News & Views: "A wrench in the motor" |
|
34. Almasy KM, Davies JP, Plate L. Comparative host interactomes of the SARS-CoV-2 nonstructural protein 3 and human coronavirus homologs. Molecular & Cellular Proteomics. 2021 (20): 100120; doi: 10.1016/j.mcpro.2021.100120
|
33. Marinko JT, Wright MT, Schlebach JP, Clowes KR, Heintzman DR, Plate L, Sanders CR. Glycosylation Limits Forward Trafficking of the Tetraspan Membrane Protein PMP22. Journal of Biological Chemistry. 2021 (296): 100719; doi: 10.1016/j.jbc.2021.100719
32. Rozas P, Pinto C, Martínez Traub F, Díaz R, Pérez V, Becerra D, Ojeda P, Ojeda J, Wright MT, Mella J, Plate L, Henríquez JP, Hetz C, Medinas DB. Protein disulfide isomerase ERp57 protects early muscle denervation in experimental ALS. Acta Neuropathologica Communications. 2021 (9): 21; doi: 10.1186/s40478-020-01116-z
|
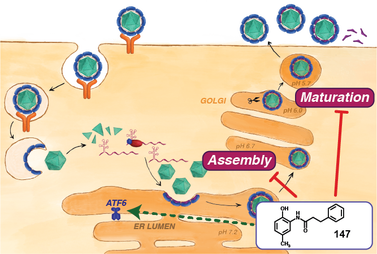
31. Almasy KM, Davies JP, Lisy SM, Tirgar R, Tran SC, Plate L. Dengue and Zika virus infection are impaired by small molecule ER proteostasis regulator 147 in an ATF6-independent manner. PNAS. 2020 118(3): e2012209118; doi: 10.1073/pnas.2012209118
Article featured in Research News @ Vanderbilt: "Newly discovered molecule disrupts virus infections through protein quality control pathways" |
30. Wright MT, Plate L. Revealing Functional Insights into ER Proteostasis through Proteomics and Interactomics. Experimental Cell Research 2021 (399): 112417; doi: 10.1016/j.yexcr.2020.112417
|
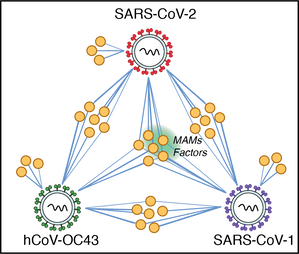
29. Davies JP, Almasy KM, McDonald JF, Plate L. Comparative multiplexed interactomics of SARS-CoV-2 and homologous coronavirus non-structural proteins identifies unique and shared host-cell dependencies. ACS Infectious Diseases 2020 (6): 3174; doi: 10.1021/acsinfecdis.0c00500
Article featured in Research News @ Vanderbilt.: "To understand how SARS-CoV-2 replicates, Vanderbilt scientists look at host cell-virus interactions" |
|
28. Wright MT, Kouba L, Plate L. Thyroglobulin Interactome Profiling Uncovers Molecular Mechanisms of Thyroid Dyshormonogenesis. Molecular & Cellular Proteomics 2021 (20): 100008; doi: 10.1074/mcp.RA120.002168
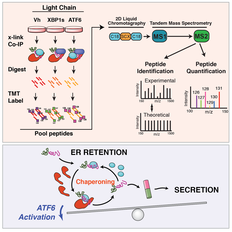
27. Vidal RL, Sepulveda D, Troncoso-Escudero P, Garcia-Huerta P, Gonzalez C, Plate L, Jerez C, Canovas J, Rivera C, Castillo V, Cisternas M, Leal S, Martinez A, Grandjean J, Lashuel H, Martin AJM , Latapiat V, Matus S, Sardi SP, Wiseman RL, Hetz C. Enforced dimerization between XBP1s and ATF6f enhances the protective effects of the unfolded protein response (UPR) in models of neurodegeneration Molecular Therapy 2021 (29): 1862; doi: 10.1016/j.ymthe.2021.01.033 |
26. Kirak O, Ke E, Yang KY, Schwartz A, Plate L, Nham A, Abadejos JR, Valencia A, Wiseman RL, Lui KO, Ku M. Premature Activation of Immune Transcription Programs in Autoimmune-Predisposed Mouse Embryonic Stem Cells and Blastocysts. International Journal of Molecular Sciences. 2020 21(16): 5743. doi: 10.3390/ijms21165743
|
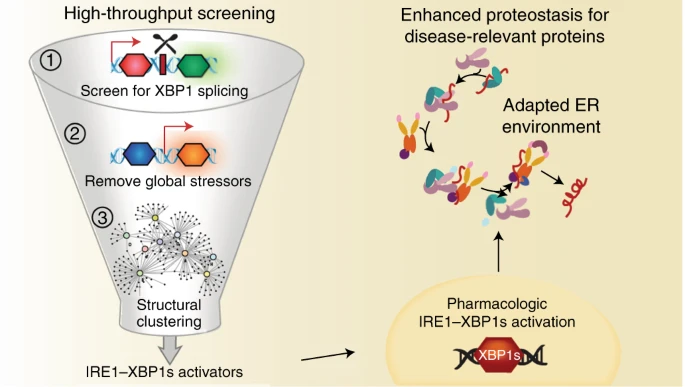
25. Grandjean JMD, Madhavan A, Cech L, Seguinot BO, Paxman, RJ, Smith E, Scampavia L, Powers ET, Cooley CB, Plate L, Spicer TP, Kelly JW, Wiseman RL. Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming. Nature Chemical Biology. 2020 (16): 1052; doi: 10.1038/s41589-020-0584-z
Article featured in Nat. Chem. Bio.: "A stress-free stress response" by Nadia Cummins and Rebecca C. Taylor. doi: 10.1038/s41589-020-0616-8 |
24. García-Huerta P, Troncoso-Escudero P, Wu D, Thiruvalluvan A, Cisternas M, Henríquez DR, Plate L, Chana-Cuevas P, Saquel C, Thielen P, Longo KA, Geddes BJ, Lederkremer GL, Sharma N, Shenkman M, Naphade S, Ellerby LM, Sardi P, Spichiger C, Richter HG, Court FA, Wiseman RL, Gonzalez-Billault C, Bergink S, Vidal RL, Hetz C. Insulin-like growth factor 2 (IGF2) protects against Huntington’s disease through the extracellular disposal of protein aggregates. Acta Neuropathologica 2020 (140): 737; doi: 10.1007/s00401-020-02183-1
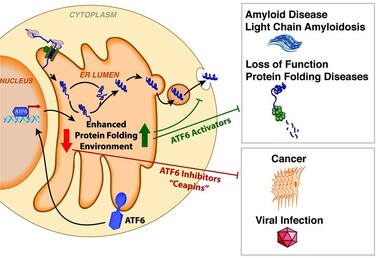
23. Torres SE, Gallagher CM, Plate L, Gupta M, Liem CR, Guo X, Tian R, Stroud RM, Kampmann M, Weissman JS, Walter P. Ceapins block the unfolded protein response sensor ATF6α by inducing a neomorphic inter-organelle tether. Elife. 2019 8: e46595 doi: 10.7554/eLife.46595
|
22. Plate, L, Rius, B, Nguyen B, Genereux JC, Kelly, JW, Wiseman, RL. Quantitative Interactome Proteomics Reveals a Molecular Basis for ATF6-Dependent Regulation of a Destabilized Amyloidogenic Protein. Cell Chemical Biology. 2019 26: 913-925. doi: https://doi.org/10.1016/j.chembiol.2019.04.001
Article featured in Cell Chemical Biology Previews: "Interactome Changes Quantified to Identify the ER Proteostasis Network to Fight Amyloid Diseases" by Ya-Juan Wang and Ting-Wei Mu. doi: https://doi.org/10.1016/j.chembiol.2019.07.003 |
21. Blackwood EA, Azizi K, Thuerauf DJ, Paxman RJ, Plate L, Kelly JW, Wiseman RL, Glembotski CC. Pharmacologic ATF6 activation confers global protection in widespread disease models by reprograming cellular proteostasis. Nat Commun. 2019 10(1):187. doi: 10.1038/s41467-018-08129-2
|
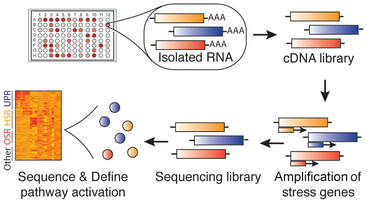
20. Grandjean JM, Plate L, Morimoto RI, Bollong MJ, Powers ET, Wiseman RL. Deconvoluting Stress-Responsive Proteostasis Signaling Pathways for Pharmacologic Activation using Targeted RNA-sequencing. ACS Chemical Biology. 2019 14(4): 784-795. doi: 10.1021/acschembio.9b00134
|
19. Paxman, RJ, Plate, L, Blackwood EA, Glembotski, CC, Powers, ET, Wiseman, RL, Kelly, JW. A Small Molecule that Preferentially Activates the ATF6 Pathway of the Unfolded Protein Response is Metabolically Activated to Selectively Modify Endoplasmic Reticulum Proteins. Elife. 2018 7: e37168. doi: 10.7554/eLife.37168
18. Kroeger H, Grimsey N, Paxman R, Chiang WC, Plate L, Jones Y, Shaw PX, Trejo J, Tsang SH, Powers E, Kelly JW, Wiseman RL, Lin JH. The unfolded protein response regulator ATF6 promotes mesodermal differentiation. Sci Signal. 2018 11(517). pii: eaan5785. doi: 10.1126/scisignal.aan5785
|
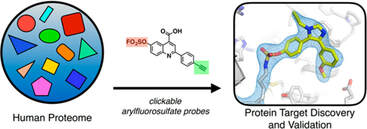
17. Mortenson DE, Brighty GJ, Plate L, Bare G, Chen W, Li S, Wang H, Cravatt BF, Forli S, Powers ET, Sharpless KB, Wilson IA, Kelly JW. “Inverse Drug Discovery” Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates. J Am Chem Soc. 2018 140(1): 200-210. doi: 10.1021/jacs.7b08366
Article featured on "In the Pipeline" (by Derek Lowe): Digging Through the Proteins, Covalently |
16. Schonhoft JD, Monteiro C, Plate L, Eisele YS, Kelly JM, Boland D, Parker CG, Cravatt BF, Teruya S, Helmke S, Maurer M, Berk J, Sekijima Y, Novais M, Coelho T, Powers ET, Kelly JW. Peptide probes detect misfolded transthyretin oligomers in plasma of hereditary amyloidosis patients. Sci Transl Med. 2017 407 pii: eaam7621. doi: 10.1126/scitranslmed.aam7621
15. Chen KC, Qu S, Chowdhury S, Noxon IC, Schonhoft JD, Plate L, Powers ET, Kelly JW, Lander GC, Wiseman RL. The endoplasmic reticulum HSP40 co-chaperone ERdj3/DNAJB11 assembles and functions as a tetramer. EMBO J. 2017 36(15). pii: e201695616. doi: 10.15252/embj.201695616.
14. Plate L, Wiseman RL. Regulating Secretory Proteostasis through the Unfolded Protein Response: From Function to Therapy. Trends Cell Biol. 2017 Jun 21. pii: S0962-8924(17)30085-5. doi: 10.1016/j.tcb.2017.05.006.
13. Alvarez-Garcia O, Matsuzaki T, Olmer M, Plate L, Kelly JW, Lotz MK. Regulated in Development and DNA Damage Response 1 Deficiency Impairs Autophagy and Mitochondrial Biogenesis in Articular Cartilage and Increases the Severity of Experimental Osteoarthritis. Arthritis Rheumatol. 2017 69 (7):1418-1428. doi: 10.1002/art.40104.
|
12. Plate L, Cooley CB, Chen JJ, Paxman RJ, Gallagher CM, Madoux F, Genereux JC, Dobbs W, Garza D, Spicer TP, Scampavia L, Brown SJ, Rosen H, Powers ET, Walter P, Hodder P, Wiseman RL, Kelly JW. Small molecule proteostasis regulators that reprogram the ER to reduce extracellular protein aggregation. Elife. 2016 Jul 20;5. pii: e15550. doi: 10.7554/eLife.15550.
11. Plate L, Paxman RJ, Wiseman RL, Kelly JW. Modulating protein quality control. Elife. 2016 Jul 20;5. pii: e18431. doi: 10.7554/eLife.18431. |
10. Chen W, Dong J, Plate L, Mortenson DE, Brighty GJ, Li S, Liu Y, Galmozzi A, Lee PS, Hulce JJ, Cravatt BF, Saez E, Powers ET, Wilson IA, Sharpless KB, Kelly
JW. Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J Am Chem Soc. 2016 Jun 15;138(23):7353-64. doi: 10.1021/jacs.6b02960.
JW. Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J Am Chem Soc. 2016 Jun 15;138(23):7353-64. doi: 10.1021/jacs.6b02960.
9. Cooley CB, Ryno LM, Plate L, Morgan GJ, Hulleman JD, Kelly JW, Wiseman RL. Unfolded protein response activation reduces secretion and extracellular aggregation of amyloidogenic immunoglobulin light chain. Proc Natl Acad Sci U S A. 2014 Sep 9;111(36):13046-51. doi: 10.1073/pnas.1406050111.
|
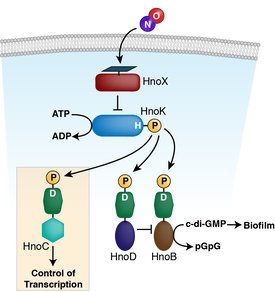
8. Plate L, Marletta MA. Phosphorylation-dependent derepression by the response regulator HnoC in the Shewanella oneidensis nitric oxide signaling network. Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):E4648-57. doi: 10.1073/pnas.1318128110.
7. Plate L, Marletta MA. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem Sci. 2013 Nov;38(11):566-75. doi: 10.1016/j.tibs.2013.08.008. 6. Plate L, Marletta MA. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol Cell. 2012 May 25;46(4):449-60. doi: 10.1016/j.molcel.2012.03.023. |
5. Ivanisevic J, Zhu ZJ, Plate L, Tautenhahn R, Chen S, O’Brien PJ, Johnson CH, Marletta MA, Patti GJ, Siuzdak G. Toward ‘omic scale metabolite profiling: a dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal Chem. 2013 Jul 16;85(14):6876-84. doi: 10.1021/ac401140h.
4. Weinert EE, Plate L, Whited CA, Olea C Jr, Marletta MA. Determinants of ligand affinity and heme reactivity in H-NOX domains. Angew Chem Int Ed Engl. 2010;49(4):720-3. doi: 10.1002/anie.200904799.
3. Carlson HK, Plate L, Price MS, Allen JJ, Shokat KM, Marletta MA. Use of a semisynthetic epitope to probe histidine kinase activity and regulation. Anal Biochem. 2010 Feb 15;397(2):139-43. doi: 10.1016/j.ab.2009.10.009.
2. Hassan AQ, Wang Y, Plate L, Stubbe J. Methodology to probe subunit interactions in ribonucleotide reductases. Biochemistry. 2008 Dec 9;47(49):13046-55. doi: 10.1021/bi8012559.
1. Gianolio DA, Philbrook M, Avila LZ, Young LE, Plate L, Santos MR, Bernasconi R, Liu H, Ahn S, Sun W, Jarrett PK, Miller RJ. Hyaluronan-tethered opioid depots: synthetic strategies and release kinetics in vitro and in vivo. Bioconjug Chem. 2008 Sep;19(9):1767-74. doi: 10.1021/bc8000479.